Lecture 1 details
- Genome Sciences 541, Jesse Bloom
- These slides are at http://jbloom.github.io/GenomeSciences541/lecture_1.html
- Reading for this lecture is
Zuckerkandl and Pauling (1965). Read pages 97-114 and 147-152.
Zuckerkandl and Pauling (1965).
Read pages 97-114 and 147-152.Zuckerkandl and Pauling (1965): one of the first molecular studies of protein evolutionWe routinely abstract the complex molecule DNA to strings of letters with little loss of relevant information:
atg gtg ctc agc gag gga gaa tgg cag ttg gtt ctg cac gtc ...
It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material. – Watson and Crick (1953)
The translation from nucleic acid to protein proceeds in a sequential fashion according to a systematic code with relatively simple rules. – Nirenberg (1968)
The properties of proteins are not easily abstracted.
Proteins are linear polymers encoded by DNA:
atg gtg ctc agc gag gga gaa tgg cag ttg gtt ctg cac gtc ...
M V L S E G E W Q L V L H V ...
Proteins derive their relevant properties from their three-dimensional structures.
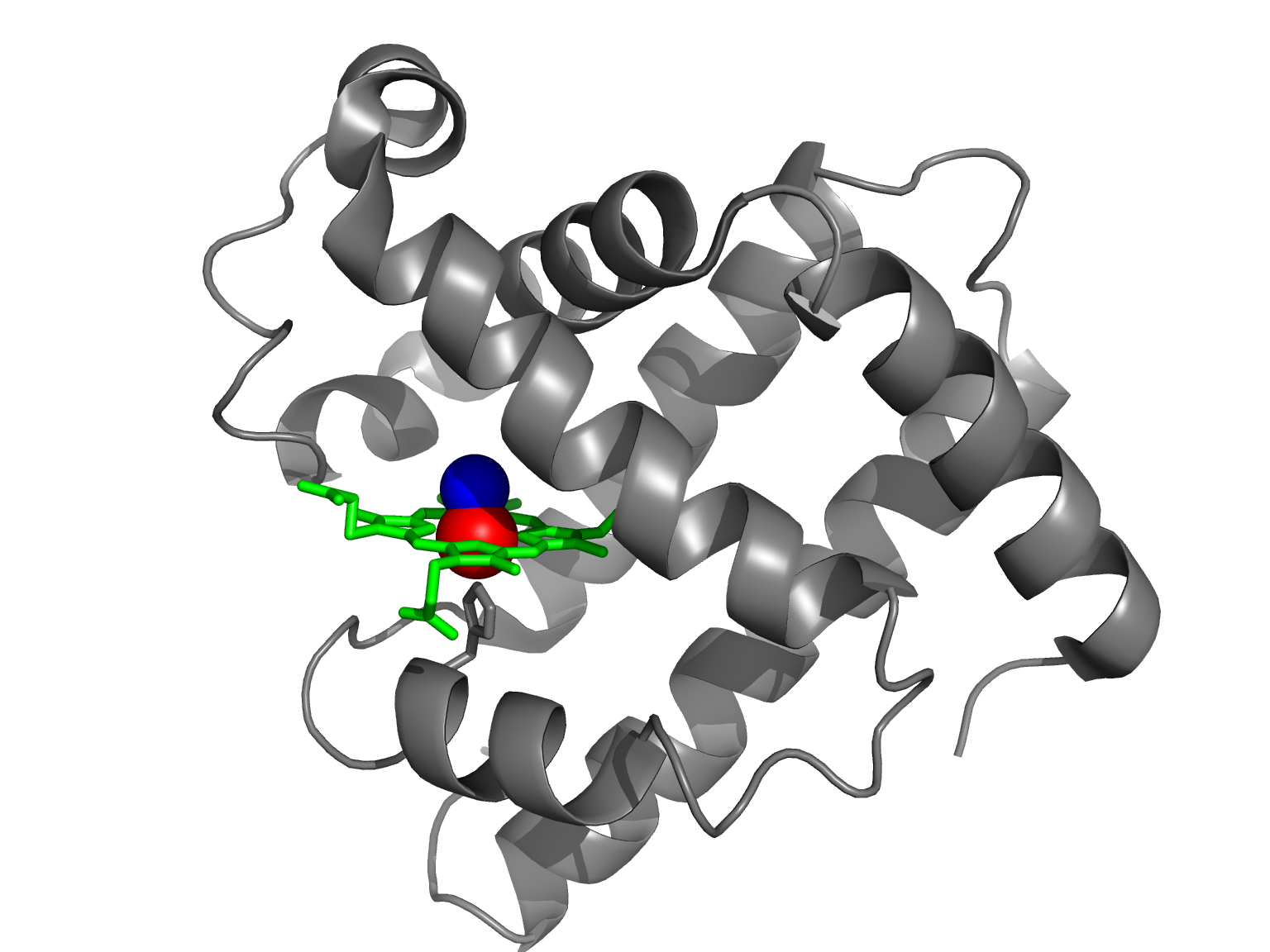
MVHLTPEEKSAVT...
MVHLTPvEKSAVT...
“Sickle Cell Anemia, a Molecular Disease” (Pauling et al, 1949)
We will perform an analysis similar to that of Zuckerkandl and Pauling, and look at some of the conclusions that they draw.
>carp
MA----DHELVLKCWGGVEADFEGTGGEVLTRLFKQHPETQKLFPKFVGIA-QSDLAGNAAVKAHGATVLKSWASCLKARGDHAAILKPLATTHANTHKIALNNFRLITEVLVKVMAEKAGLD--AGGQSALRRVMDVVIGDIDTYYKEIGFAG
>chicken
MGLSDQEWQQVLTIWGKVEADIAGHGHEVLMRLFHDHPETLDRFDKFKGLKTPDQMKGSEDLKKHGATVLTQLGKILKQKGNHESELKPLAQTHATKHKIPVKYLEFISEVIIKVIAEKHAADFGADSQAAMKKALELFRNDMASKYKEFGFQG
>horse
MGLSDGEWQQVLNVWGKVEADIAGHGQEVLIRLFTGHPETLEKFDKFKHLKTEAEMKASEDLKKHGTVVLTALGGILKKKGHHEAELKPLAQSHATKHKIPIKYLEFISDAIIHVLHSKHPGDFGADAQGAMTKALELFRNDIAAKYKELGFQG
>human
MGLSDGEWQLVLNVWGKVEADIPGHGQEVLIRLFKGHPETLEKFDKFKHLKSEDEMKASEDLKKHGATVLTALGGILKKKGHHEAEIKPLAQSHATKHKIPVKYLEFISECIIQVLQSKHPGDFGADAQGAMNKALELFRKDMASNYKELGFQG
>mouse
MGLSDGEWQLVLNVWGKVEADLAGHGQEVLIGLFKTHPETLDKFDKFKNLKSEEDMKGSEDLKKHGCTVLTALGTILKKKGQHAAEIQPLAQSHATKHKIPVKYLEFISEIIIEVLKKRHSGDFGADAQGAMSKALELFRNDIAAKYKELGFQG
>sperm-whale
MVLSEGEWQLVLHVWAKVEADVAGHGQDILIRLFKSHPETLEKFDRFKHLKTEAEMKASEDLKKHGVTVLTALGAILKKKGHHEAELKPLAQSHATKHKIPIKYLEFISEAIIHVLHSRHPGDFGADAQGAMNKALELFRKDIAAKYKELGYQG
>tuna
MA----DFDAVLKCWGPVEADYTTMGGLVLTRLFKEHPETQKLFPKFAGIA-QADIAGNAAISAHGATVLKKLGELLKAKGSHAAILKPLANSHATKHKIPINNFKLISEVLVKVMHEKAGLD--AGGQTALRNVMGIIIADLEANYKELGFSG
>turtle
MGLSDDEWNHVLGIWAKVEPDLTAHGQEVIIRLFQLHPETQERFAKFKNLTTIDALKSSEEVKKHGTTVLTALGRILKQKNNHEQELKPLAESHATKHKIPVKYLEFICEIIVKVIAEKHPSDFGADSQAAMKKALELFRNDMASKYKEFGFLG
>zebrafish
MA----DHDLVLKCWGAVEADYAANGGEVLNRLFKEYPDTLKLFPKFSGIS-QGDLAGSPAVAAHGATVLKKLGELLKAKGDHAALLKPLANTHANIHKVALNNFRLITEVLVKVMAEKAGLD--AAGQGALRRVMDAVIGDIDGYYKEIGFAG
Sequences at Analysis of myoglobin homologs
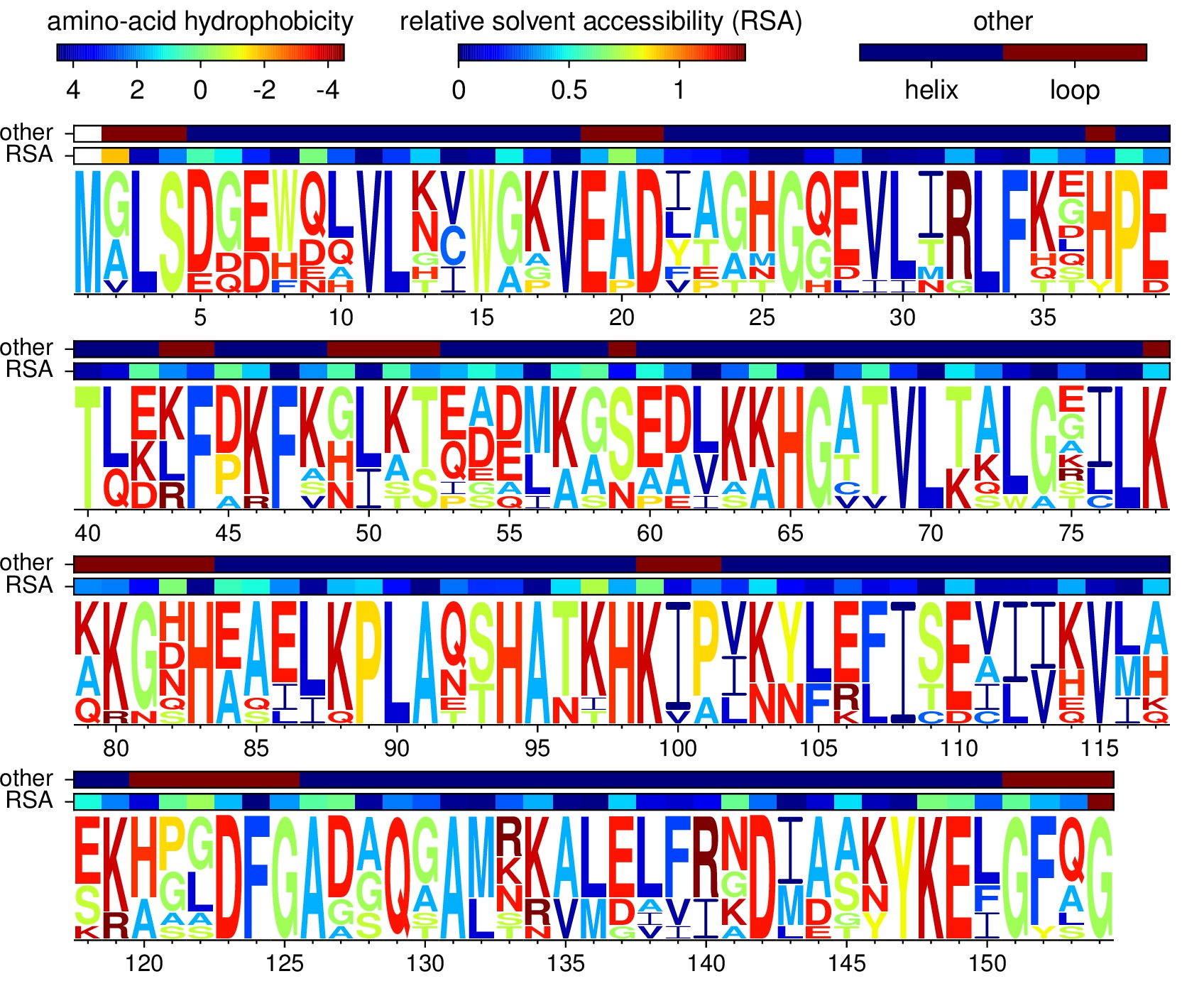
Code /data to produce this figure at Analysis of myoglobin homologs
Deletions or additions of one to several amino acid residues are expected to be eliminated by natural selection in a high proportion of cases. Those that are preserved should be mostly found at either end of a chain, at the end of helices, in short helices, or in nonhelical regions, notably in loops that may be shortened or lengthened without affecting the steric relationships in the rest of the molecule. A deletion or addition in the middle of a long helix would result in so many simultaneous alterations in side-chain interactions that it is highly unlikely that the tertiary structure and the function of the molecule could survive such an event. The deletions or additions found in hemoglobin and myoglobin chains are compatible with these generalities.
This observation has stood the test of time. In fact, sequences with as little as 30-35% identity generally have very similar structures. See Chothia and Lesk (1984) and Sander and Schneider (1991).
Many such substitutions may lead to relatively little fuinctional change, whereas at other times the replacement of one single amino acid residue by another may lead to a radical functional change… Of course, the two aspects are not unrelated, since the functional effect of a given single substitution will frequently depend on the presence or absence of a number of other substitutions.
What do we call that phenomenon?
The extent of sequence change can be used to reconstruct evolutionary relationships (molecular phylogenetics)
However, the constant background sequence change can make it difficult to pinpoint biologically important changes:
- Positive selection : for instance, take a look at nextflu
- Disease-causing mutations